|
 |
| Korean J Ophthalmol > Volume 28(4); 2014 > Article |
Abstract
Purpose
To investigate the relationship between plasma TDRD7 mRNA and lens transparency, and to evaluate plasma TDRD7 mRNA as a potential marker for cataracts and its sub-type by quantitatively analyzing human peripheral blood.
Methods
Plasma RNA was extracted from 40 patients with cataracts, and 30 normal controls of matched age and gender. Blood cholesterol and fasting glucose were measured, and the RNA extracted from the sample was synthesized into cDNA. After polymerase chain reaction, the results were compared after quantifying the TDRD7 mRNA using ABL1 mRNA for normalization. We analyzed the relative gene expression data via the ΔΔCt method.
Results
The normalized 2-ΔΔCt of plasma TDRD7 mRNA based on ABL1 mRNA was 1.52 ± 0.63 in the case of the control group and 1.05 ± 0.34 in the case of the cataract patients, and the TDRD7 expression level of the cataract patients was lower than that of the control group (p = 0.048). The comparison of the genetic values of different types of cataracts demonstrated that the TDRD7 expression level of the cortical type and mixed type were lower than those of the nuclear type and posterior subcapsular opacity type (p = 0.017).
Cataract is globally the primary cause of blindness, and is the cause of blindness in approximately 15 million people, accounting for more than half of the blind. It evidences a prevalence rate of 24.1% of adults over the age of 19% and 93.7% of adults over the age of 70 in Korea [1]. According to a previous report, cataracts occur in 32.8% to 43.5% of adults over the age of 40, and the prevalence rate noticeably increases with advancing age; initial signs of cataract are seen in almost 100% of elderly individuals in their 80s [2]. The factors which are thus far known to directly influence the occurrence of cataract include ocular trauma, intraocular surgery, endophthalmitis, long-term use of steroids, and congenital cataracts [3]. However, the causes of most cases of cataracts have not been found, although extensive research has identified the risk factors for cataracts to be systemic conditions such as age, gender, smoking status, myopia, and diabetes, as well as ultraviolet irradiation [4,5,6,7].
Recently, gene-related research has been extensively conducted in the fields of medicine and biology, thanks to advancements in genetic engineering, and in ophthalmology as well, gene-related research has been conducted into retinal vascular diseases, age-related macular degeneration, glaucoma, and cataracts [8,9,10]. While cataracts represent a common end stage of mutations in a potentially large number of genes acting via varied mechanisms, in practice most inherited cataracts are associated with a subgroup of genes encoding for proteins of particular importance in the maintenance of lens transparency and homeostasis [8,9,10]. The increasing availability of more detailed information regarding these proteins and their functions is making it possible to understand the pathophysiology of cataracts and the biology of the lens in general. Recently, Lachke et al. [11] determined that RNA granules, which play a key role in messenger RNA (mRNA) processing, can affect eye development, leading to juvenile cataracts in humans and mice. They also verified a malfunctioning gene, TDRD7, in a mouse strain that develops cataracts. TDRD7 is a scaffold protein, the specific function of which is unknown. It has been identified in complexes with proteins regulating cytoskeletal dynamics, movement of the cytocentrum, and the transportation of mRNA and organs which translate proteins. TDRD7 encodes for a component of the cytoplasmic RNA granules which are involved in determining the fate of mRNAs. The component of specific cytoplasmic RNA granules involved in the post-transcriptional regulation of specific genes most probably acts by binding to specific mRNAs and regulating their translation. If TDRD7 fails to build an essential protein, the lens cannot develop normally. Lachke et al. [11] discovered the absence of this protein in some children, as well as a type of structure known as RNA granules in the lenses of mice. RNA granules serve to regulate mRNAs in the cell. The mRNA's primary job is to serve as a template to carry DNA-encoded information from the nucleus into the cytoplasm or body of the cell, providing blueprints for protein production. TDRD7 loss-of-function mutations affect the regulation of mRNA, and as a result, loss-of-function mutations have been implicated in cataract causation. The results of this study suggest that RNA granules are important in TDRD7 for lens transparency.
The subjects of the present study were 40 patients with cataracts and 30 normal persons who visited the Department of Ophthalmology. The contents of the survey and examination were explained to all of the subjects, and written consent was obtained. The questionnaire assessed gender, age, and life habit variables such as smoking status, drinking status, and daily life, in addition to survey items about personal history. The medical examinations included measurements of height, weight, blood pressure, fasting plasma glucose, and total cholesterol and genetic tests for ABL1 and TDRD7. For the genetic and clinical examinations involving survey, we obtained approval from the institutional review board of Chosun University Hospital. Both groups belonged to the same ethnicity. Thirty healthy, age- and sex-matched subjects were also included for the controls. Blood for testing was obtained before noon the next day after instructing the subjects to keep their stomachs empty for 12 hours or longer after eating dinner. For serum cholesterol, plasma glucose and gene analysis, peripheral blood (4-5 mL) was taken into vacuum tubes treated with EDTA (ethylene diamine tetraacetic acid), and transported on ice from the time it was obtained until centrifugation. The blood that arrived at the test room was then centrifuged for 10 minutes at 3,000 rpm at 4Ōäā prior to testing. Total cholesterol and plasma glucose were measured using a commercial analytical system (Architect ci16200 Analyser; Abbott Laboratories, Abbott Park, IL, USA).
Total RNA was extracted from each venous blood sample with the EasyRed RNA extraction kit (Intronbio, Seoul, Korea) in accordance with the manufacturer's protocol. The cDNA synthesis was conducted using a Superscript VILO cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol. For quantification of the TDRD7 transcript relative to the ABL1 control gene, we used the primers specific for TDRD7 (forward: 5'-ACGAGTAGAGATCACAAATG-3'; reverse: 5'-TCTGCTAAACAGCACTTAAT-3') and ABL1 (forward 5'-CTGCTAGAGAAGGACTACC-3'; reverse: 5'-TCGTCTGAGATACTGGATTC-3'). Real-time polymerase chain reaction (PCR) reactions with the above primers were conducted in a total volume of 20 ┬ĄL using Phusion Flash (Thermo Scientific, Wilmington, DE, USA) with EvaGreen (Biotium, Hayward, CA, USA) in accordance with the manufacturer's recommendations. Thermocycling was carried out using a CFX96 real-time PCR detection system (Bio-rad, Hercules, CA, USA) with the following conditions: 1 cycle of 98Ōäā for 10 seconds, 40 cycles of 98Ōäā for 1 second, and 55Ōäā for 15 seconds. The amplification curves of 2 genes, TDRD7 and ABL1 mRNA, are shown in Fig. 1.
Amplification of an endogenous control may be conducted to standardize the amount of TDRD7. For the quantification of gene expression, we employed ABL1 mRNA as the endogenous control. The relative expression levels of TDRD7 mRNA, normalized to ABL1 mRNA, were calculated using the 2-ΔΔCt value. In order to apply the 2-ΔΔCt method, the results of real-time reverse transcription-PCRs were represented as cycle threshold (Ct) values. The Ct value was defined as the cycle at which a sample crosses a threshold that is significantly above the background fluorescence and within the exponential phase of the amplification. The average of three Ct measurements was calculated for both the TDRD7 and ABL1 mRNA. ΔCt was determined as the mean of the triplicate Ct values for the TDRD7 genes minus the mean of the triplicate Ct values for the ABL1 gene. The ΔΔCt represented the difference between the two groups for a given target gene, namely ΔΔCt = ΔCt (cataract group) - ΔCt (the mean of normal group). The x-fold higher expression of a given target gene in the cataract group compared to the normal group was calculated as 2-ΔΔCt. All assays were executed in triplicate.
A past history of ophthalmologic disease such as ocular trauma and cataract surgery was checked, as well as whether the subjects had systemic disease such as hypertension, diabetes, or cardiovascular disease, along with family history. In addition to uncorrected visual acuity, corrected visual acuity was examined in the case of subjects that wore corrective lenses, and the best corrected visual acuity was measured after checking the refractive state via a manifest refraction test. The state of the lens was classified by Lens Opacities Classification System (LOCS) III through an examination using a slit lamp microscope after mydriasis [12]. If, in one eye or two eyes, there was nuclear opacity of second stage or higher in LOCS III classification, cortical opacity of second stage or higher, or posterior subcapsular opacity of first stage or higher, those subjects were classified as cataract patients, and were otherwise categorized as control group members. Cataract patients were classified into nuclear, cortical, posterior subcapsular, or mixed types. Cases of pseudophakic eye after cataract surgery were excluded from the classifications.
Data analysis was conducted using SPSS ver. 17.0 (Chicago, IL, USA) and the actual numbers, percentage, average, and standard deviation were obtained as descriptive statistics in order to study the general characteristics of the patient group and control group. The comparison of the general characteristics of the patient group and the control group was conducted via t test and Žć2 test, and, in comparison of the genetic values of the patient group and the control group, cases in which the p-value was found to be below 0.05 by an independent sample t test were determined to be statistically significant. In order to compare the TDRD7 gene values of different types of cataracts, one-way ANOVA was used; to compare the TDRD7 gene values of normal eyes of different age objects, correlation analysis was conducted. Cases in which the p-value was below 0.05 were determined to be statistically significant.
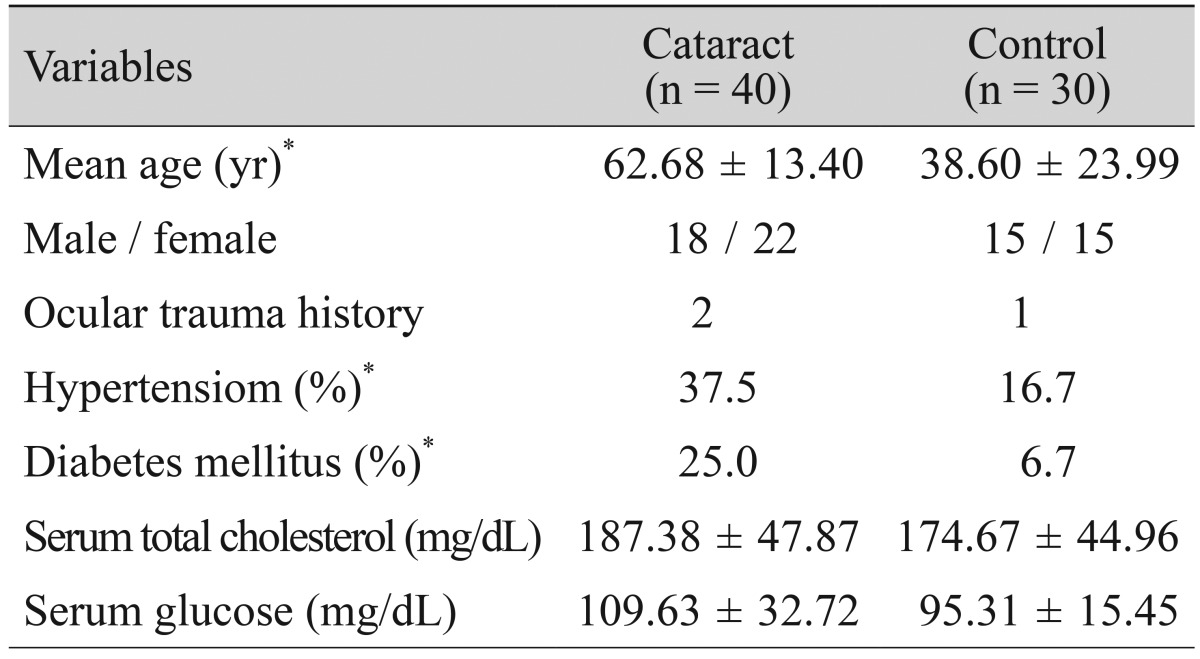
Among the total 70 subjects, 40 (57.1%) were cataract patients while 30 (42.9%) belonged to the control group; 33 (47.1%) were males and 37 (52.9%) were females. The average age of the whole subject population was 52.67 ┬▒ 12.96 years and the age range of the subjects was between 10 and 89 years. The age of the cataract patients was significantly higher, with an average of 62.68 ┬▒ 13.40 years, whereas the average age of the control group was 38.60 ┬▒ 23.99 years (p < 0.001). The frequencies of medical history of hypertension and diabetes of the cataract patients were higher than those of the control group, evidencing values of 37.5% and 25.0%, respectively; the differences between these values were statistically significant (p < 0.05). Although the total cholesterol of the cataract patients was higher than that of the control group with a value of 187.38 ┬▒ 47.87 mg/dL, while that of the control group was 174.67 ┬▒ 44.96 mg/dL, the difference was not statistically significant (p > 0.05). Although the serum glucose level of the cataract patients was higher than that of the control group with a value of 109.63 ┬▒ 32.72 mg/dL, while that of the control group was 95.31 ┬▒ 15.45 mg/dL, the difference was not statistically significant (p > 0.05) (Table 1). In the results of our classification of the cataracts of 40 patients, in accordance with the positions of the opacity, the cataracts were identified as 19 nuclear types, 10 cortical, 6 posterior subcapsular, and 5 mixed types.
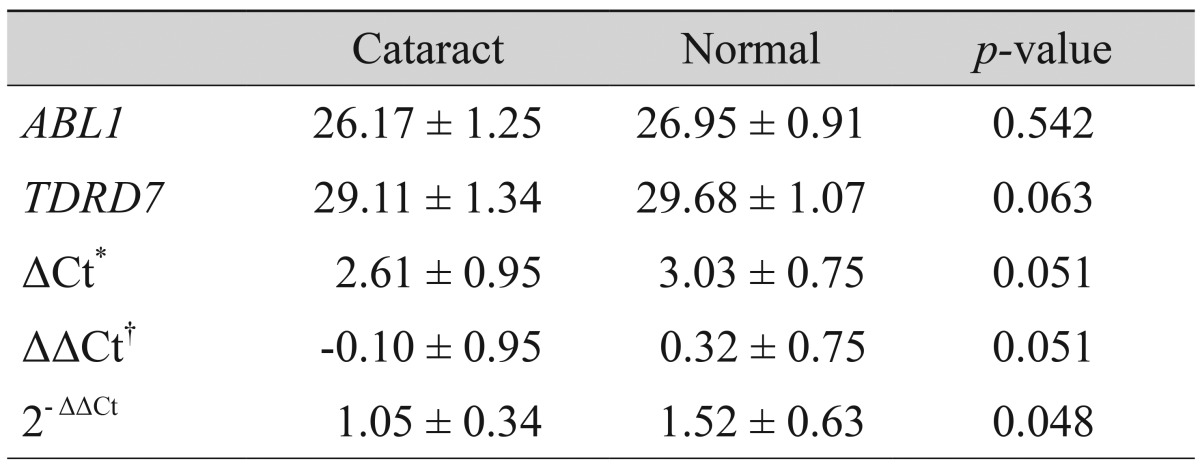
The ABL1 and TDRD7 values of the control group were 26.95 ± 0.91 and 29.68 ± 1.07, respectively, and those of the cataract patients were 26.17 ± 1.25 and 29.11 ± 1.34, respectively. ΔCt was determined as the mean of the triplicate Ct values for the TDRD7 genes minus the mean of the triplicated Ct values for the ABL1 gene. ΔΔCt represented the difference between the two groups for a TDRD7 gene, more precisely ΔΔCt = ΔCt (cataract group) - ΔCt (normal group). The mean and standard deviations for all ΔCt and ΔΔCt values (cataract versus normal group) are provided in Table 2. The actual expression level of a TDRD7 gene was analyzed using the 2-ΔΔCt value. The results, including standard deviations, are listed in Table 2. As the result of adjusting the TDRD7 value in reference to ABL1 mRNA, the TDRD7 expression level of the cataract patients was found to be significantly lower than that of the control group, with a value of 1.05 ± 0.34, whereas that of the control group was 1.52 ± 0.63 (p = 0.048) (Table 2). To allow for a better overview and to show the broad spectrum of expression rates, all 2-ΔΔCt values (with standard deviations) are illustrated within a single figure (Fig. 2).
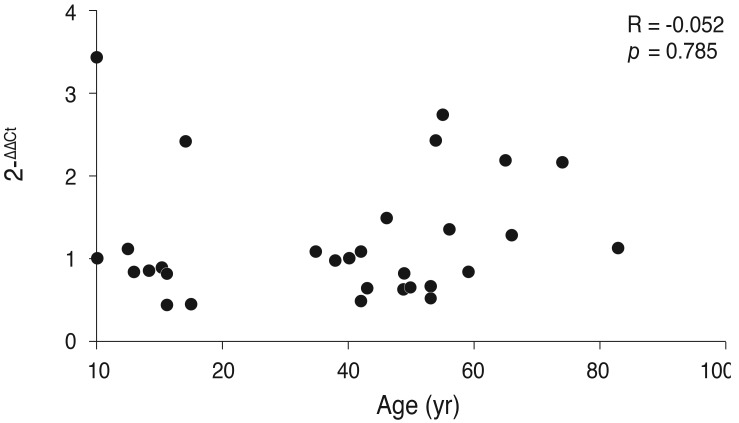
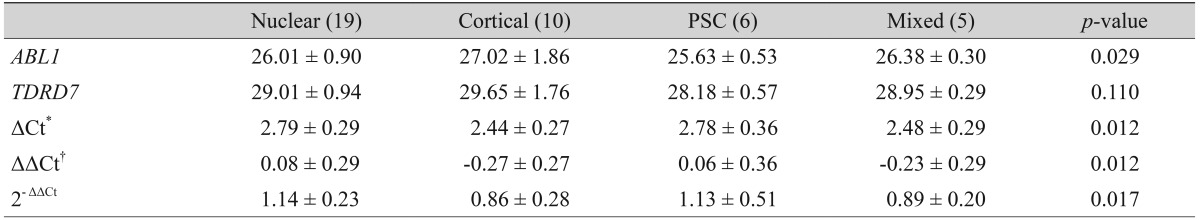
The results of our comparisons of the genetic values of different types of cataracts demonstrated that the TDRD7 expression levels of the cortical type and mixed type were statistically significantly lower than those of the nuclear type and posterior subcapsular opacity type (p = 0.017) (Table 3). To allow for a better overview and to demonstrate the wide spectrum of expression rates, all 2-ΔΔCt values (with standard deviations) are illustrated within a single figure (Fig. 3). The results of measuring the TDRD7 gene expression levels of the normal eyes of different age objects demonstrated that there was no statistically significant correlation between the two variables (p = 0.785) (Fig. 4).
This study focused on the loss-of-function mutation of TDRD7, which has been shown to affect its expression level and the clinical presentation of cataracts. Cataracts account for approximately 50% of blindness globally, and the only way to treat cataracts is to remove the lens through surgery. Cataract surgery has become the most frequently executed surgery globally among the aged [13]. Understanding the causes of cataracts and risk factors can be helpful in the search for non-operative methods to delay the occurrence of or prevent cataracts. Most known gene mutations that lead to congenital cataracts derive from genes that encode for structural proteins of the lens. The gene mutations involved in the generation of transcription factors that regulate the expression of structural genes account for the subset of mutations that cause cataracts. There are currently several reported mutations in congenital and hereditary cataracts involving genes that encode proteins with structural, metabolic, and transport functions [14]. It is possible that some of these genes may be involved in adult cataract with late phenotype expression due to the interaction with several harmful factors along with the aging process.
Lachke et al. [11] suggested that RNA-containing granules can regulate the subcellular localization of mRNA as well as the processing which plays an important role in the transparency of the lens. Cytoplasmic RNA granules play a role in determining whether mRNAs undergo degradation, stabilization, or intracellular localization. The structure of the lens in vertebrates requires a very high concentration of specialized cytoplasmic proteins: the crystalline, a specialized intermediate filament cytoskeleton to stabilize the structure of the lens [15]; cellular membranes with high concentrations of both the lens-specific water channel, aquaporin 0; and lens-preferred connexins, which allow the lens to transport nutrients and waste products despite its lack of blood vessels [16]. A number of the genes encoding for these proteins are abundantly expressed in the lens, resulting in a highly biased transcriptome [17]. Lachke et al. [11] have reported the identification of Tudor domain-containing 7 protein, or TDRD7, as an RNA granule component with a highly enriched and conserved pattern of developmental expression in the ocular lens. They suggested that human TDRD7 mutations result in cataract formation via the misregulation of specific, developmentally critical lens transcripts.
The official name of the TDRD7 gene is "tudor domain containing 7"; it is also referred to by other names such as CATC4, KIAA1529, PCTAIRE2BP, RP11-508D10.1, and TRAP. It is located on the long(q) arm of chromosome 9 at position 22.33. More accurately, it is located from base pair 100,174,301 to base pair 100,258,406 on chromosome 9. TDRD7 is a member of a large family of Tudor domain-containing proteins that mutually interact with methylated arginine residues on other proteins. Many members of the TDRD family, including TDRD7, are found in RNA granules-cytoplasmic complexes of protein and RNA that regulate gene expression by influencing RNA degradation, stabilization, and the intracellular localization of RNA [18]. Although TDRD7 transcripts and proteins are found in a variety of cells, especially of the male germ line, TDRD7 expression is very high in lens fiber cells and may control the production of extremely high amounts of structural proteins of the lens. TDRD7 is a Tudor domain RNA binding protein which is expressed in lens fiber cells, and is required for the posttranscriptional control of mRNAs that are essential for normal lens development and RNA granule function. Lachke et al. [11] demonstrated that TDRD7 is found in a granular distribution in lens fiber cells and colocalizes with concentrations of RNA found in the cytoplasm of these cells. They demonstrated that human organogenesis defects can result from the perturbation of a distinct, tissue-specific RNA granule component that regulates the posttranscriptional steps of developmentally critical mRNAs. Simple RNA distribution within lens fibers would presumably be inefficient because of the relatively long distance between the lens fiber cell nucleus and the fiber cell tips, as well as the high concentration of proteins in the fiber cell cytoplasm [19]. Accordingly, when we look into the matter retrospectively, the requirement for a lens-preferred RNA transport mechanism may not be entirely surprising. Most notably, the lens epithelial cells express mRNAs for lens structural proteins that are not translated into proteins until the onset of fiber cell differentiation, which is also when the onset of TDRD7 expression occurs [20]. Additionally, another possible role for TDRD7-containing RNA granules would be to regulate the translation of fiber cell structural proteins. Lachke et al. [11] determined that reduced TDRD7 expression in a lens epithelial cell line induced statistically significant changes in the expression levels of 6% of the genes expressed in those cells, including some crystallins. Overall, Lachke et al. [11] demonstrated that the vertebrate lens contains several classes of RNA granules and that TDRD7-containing granules are critically important for the normal functioning of the lens. These findings open up a new field of investigation that may ultimately help to elucidate both the physiological and biochemical functions that regulate RNA granules in the lens.
When we look into different risk factors of cataract, there are some risk factors which clearly affect specific types of cataracts. For example, smoking increases the prevalence of nuclear cataracts, and steroids are related to the occurrence of posterior subcapsular cataract [5]. The results of comparing the gene values of different types of cataracts in this study demonstrated that the TDRD7 expression ratios of the cortical type and mixed type were lower than those of the nuclear type and posterior subcapsular type; this implies that, while many senile cataracts are of nuclear type, many cataracts caused by genetic loss-of-function mutations may be of the cortical type.
Although blood cholesterol is not known as a risk factor for cataracts, a case was recently reported wherein posterior subcapsular cataract occurred in two eyes of a lathosterolosis patient showing a defect of cholesterol biosynthesis [21]; Klein et al. [22] suggested that the ingestion of statin, which is widely used for the purpose of preventing cardiovascular disease by lowing the blood cholesterol value lowers the incidence rate of nuclear cataracts. In this study, although the total cholesterol value of cataract patients was higher than that of the control group, this was not statistically significant. The result of comparing the age and the TDRD7 gene expression ratios of the patients with normal eyes demonstrated that there is no statistically significant correlation between the two variables (Fig. 4).
Although this study is significant in that it is the first study regarding the relationship between the TDRD7 gene and cataracts using Koreans as subjects, further proactive studies with a greater number of subjects selected from a larger number of regions are needed to more clearly establish the relationship between the TDRD7 gene and cataracts.
In conclusion, as the TDRD7 gene expression levels of the cataract patients found in a test conducted with 70 subjects living in a local region in the present study were statistically significantly lower than those of the control group, we can assert that there is a causal relationship between the occurrence of human cataracts and the loss- of-function mutations of the TDRD7 gene.
REFERENCES
1. Yoon KC, Mun GH, Kim SD, et al. Prevalence of eye diseases in South Korea: data from the Korea National Health and Nutrition Examination Survey 2008-2009. Korean J Ophthalmol 2011;25:421-433.



2. Shyn KH, Kim JC, Kim WS, et al. An epidemiological study of the risk factors contributing to the senile cataractogenesis by the Korean Cooperative Cataract Epidemiology Study Group (KCCESG). J Korean Ophthalmol Soc 1992;33:127-134.
3. Hodge WG, Whitcher JP, Satariano W. Risk factors for age-related cataracts. Epidemiol Rev 1995;17:336-346.


4. Ederer F, Hiller R, Taylor HR. Senile lens changes and diabetes in two population studies. Am J Ophthalmol 1981;91:381-395.


5. Delcourt C, Cristol JP, Tessier F, et al. Risk factors for cortical, nuclear, and posterior subcapsular cataracts: the POLA study. Pathologies Oculaires Liees a l'Age. Am J Epidemiol 2000;151:497-504.


6. West SK, Valmadrid CT. Epidemiology of risk factors for age-related cataract. Surv Ophthalmol 1995;39:323-334.


7. Leske MC, Chylack LT Jr, Wu SY. The lens opacities case-control study. Risk factors for cataract. Arch Ophthalmol 1991;109:244-251.


8. Zetterberg M, Tasa G, Prince JA, et al. Methylenetetrahydrofolate reductase genetic polymorphisms in patients with cataract. Am J Ophthalmol 2005;140:932-934.


9. Utheim OA, Ritland JS, Utheim TP, et al. Apolipoprotein E genotype and risk for development of cataract and age-related macular degeneration. Acta Ophthalmol 2008;86:401-403.


10. Zetterberg M, Zetterberg H, Palmer M, et al. Apolipoprotein E polymorphism in patients with cataract. Br J Ophthalmol 2004;88:716-718.



11. Lachke SA, Alkuraya FS, Kneeland SC, et al. Mutation in the RNA granule component TDRD7 cause cataract and glaucoma. Science 2011;331:1571-1576.



12. Chylack LT Jr, Wolfe JK, Singer DM, et al. The Longitudinal Study of Cataract Study Group. The lens opacities classification system III. Arch Ophthalmol 1993;111:831-836.


13. Steinberg EP, Javitt JC, Sharkey PD, et al. The content and cost of cataract surgery. Arch Ophthalmol 1993;111:1041-1049.


15. Song S, Landsbury A, Dahm R, et al. Functions of the intermediate filament cytoskeleton in the eye lens. J Clin Invest 2009;119:1837-1848.



16. Donaldson P, Kistler J, Mathias RT. Molecular solutions to mammalian lens transparency. News Physiol Sci 2001;16:118-123.


17. Wistow G, Bernstein SL, Wyatt MK, et al. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis 2002;8:171-184.

18. Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009;10:430-436.


19. Fagerholm PP, Philipson BT, Lindstrom B. Normal human lens: the distribution of protein. Exp Eye Res 1981;33:615-620.


20. Wang X, Garcia CM, Shui YB, Beebe DC. Expression and regulation of alpha-, beta-, and gamma-crystallins in mammalian lens epithelial cells. Invest Ophthalmol Vis Sci 2004;45:3608-3619.


Fig.┬Ā1
Comparison of gene expression of ABL1 and TDRD7 genes in cataract group versus normal group by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). Amplification curves for A) ABL1, B) TDRD7 (normal group), and C) TDRD7 (cataract group). The threshold level is given by a horizontal line. The cycle at which the mean amplification curve of ABL1 or TDRD7 real-time RT-PCRs crosses the threshold (cycle threshold value) is indicated by a vertical line. RFU = relative fluorescence units.
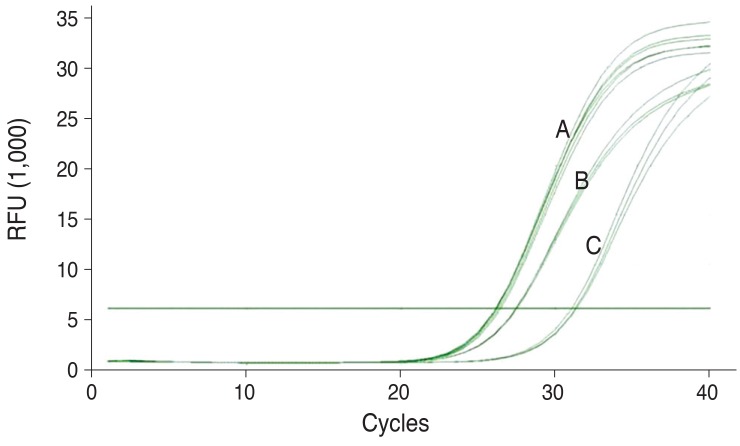
Fig.┬Ā2
Normalized plasma TDRD7 mRNA in the patient group amount relative to that in normal control group. Fold change was expressed as 2-ΔΔCt (ΔCt [TDRD7-ABL1] of each case-average ΔCt [average TDRD7 Ct-average ABL1 Ct] of normal control group). Ct = cycle threshold.
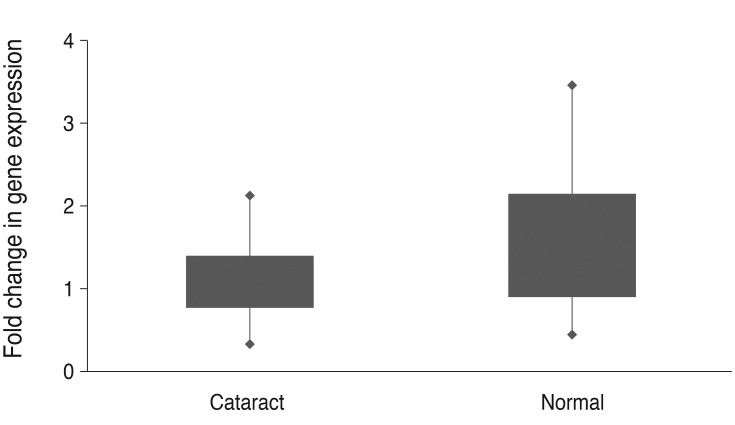
Fig.┬Ā3
Visualization for the comparison of the 2-ΔΔCt value according to the type of cataract. Fold change was expressed as 2-ΔΔCt (ΔCt [TDRD7-ABL1] of each case-average ΔCt [average TDRD7 Ct-average ABL1 Ct] of normal control group). PSC = posterior subcapsular; Ct = cycle threshold.
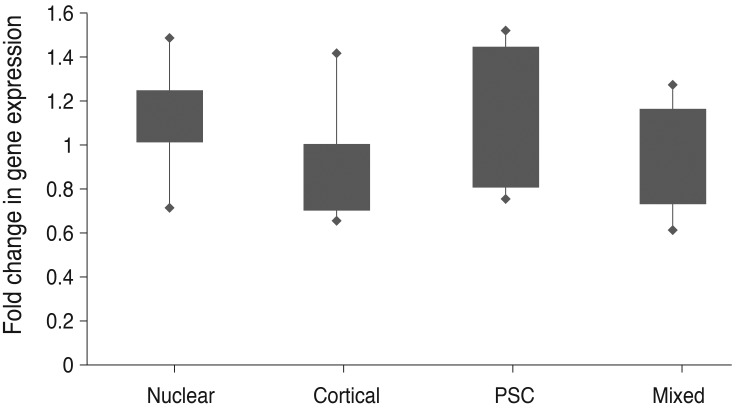
Fig.┬Ā4
Correlation analysis between age and TDRD7 gene expression levels in the blood of the normal group by polymerase chain reaction.
Table┬Ā2
Relative quantitation using the comparative Ct method in the cataract group and the normal group
- TOOLS



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print